NG-Tax 2.0 allows FAIR high-throughput analysis and classification of marker gene amplicon sequences. In this article, we describe the performance of NG-Tax 2.0. Moreover, we demonstrate its use with examplary data from the DIABIMMUNE project.
By Jasper Koehorst / July 7, 2021
KEY MESSAGES
- NG-Tax 2.0 is a semantic framework for FAIR high-throughput analysis and classification of marker gene amplicon sequences.
- NG-Tax 2.0 showed higher precision than QIIME2-DADA2, while recall was comparable.
- Next, using data from the DIABIMMUNE project, we show examples of possible data analysis
NG-Tax 2.0 is a semantic framework for FAIR high-throughput analysis and classification of marker gene amplicon sequences. These include bacterial and archaeal 16S ribosomal RNA (rRNA), as well as eukaryotic 18S rRNA and ribosomal intergenic transcribed spacer sequences. The framework can directly use single or merged reads, paired-end reads and unmerged paired-end reads from long range fragments as input. From these, it generates de novo Amplicon Sequence Variants (ASV). Subsequently, using the RDF data model, ASVs can be automatically stored in a graph database as objects that link ASV sequences with the full data-wise and element-wise provenance. Hereby, it achieves the level of interoperability required to utilize such data to its full potential.
Analysis of the data
Subsequently, the graph database can be directly queried, allowing for comparative analyses of over thousands of samples. It is, moreover, connected with an interactive Rshiny toolbox for analysis and visualization of (meta)data. Additionally, NG-Tax 2.0 exports an extended BIOM 1.0 (JSON) file as starting point for further analyses by other means. This file contains new attribute types to include information about the command arguments used, the sequences of the ASVs formed as well as classification confidence scores. Finally, it is backwards compatible. In summary, the figure below describes the whole NG-Tax 2.0 workflow.
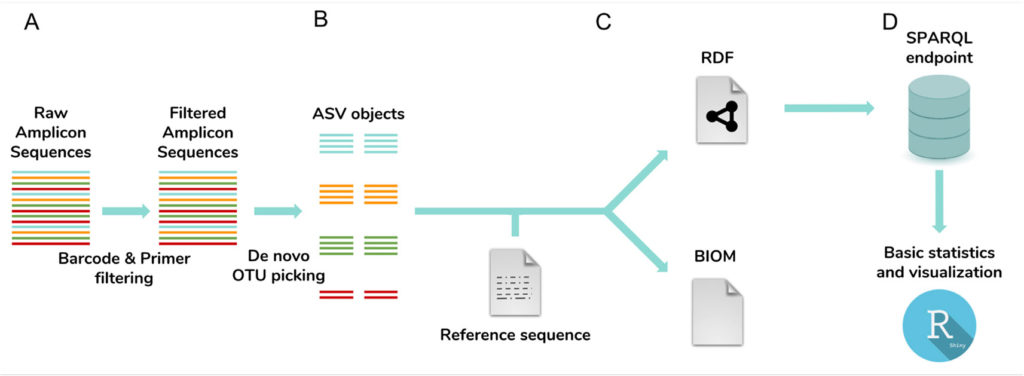
Performance and availability
We compared the performance of NG-Tax 2.0 with DADA2 using the plugin in the QIIME 2 analysis pipeline. To this aim, we obtained and evaluated fourteen 16S rRNA gene amplicon mock community samples from the literature. As a result, precision of NG-Tax 2.0 turned out to be significantly higher with an average of 0.95 vs 0.58 for QIIME2-DADA2. Meanwhile, recall was comparable with an average of 0.85 and 0.77, respectively. NG-Tax 2.0 is written in Java. Under the MIT License, you can freely access the code, ontology, a Galaxy platform implementation, the analysis toolbox, as well as tutorials and example SPARQL queries.
Working example: DIABIMMUNE project
As a working example, we used existing raw 16S rRNA gene data from the DIABIMMUNE Microbiome project and NG-Tax 2.0 for data analyses. To this end, we downloaded raw amplicon data of over 1800 microbial samples from the project. Next, the data was automatically ingested by the UNLOCK infrastructure and stored according to the ISA standard. We also captured the metadata of these samples. Then, NG-Tax 20 automatically analysed the amplicon data to generate ASVs as we described above and exemplified below. The queries used to generate the examples shown can also be embedded in or be part of standard operating procedures (SOPs). Further post-processing can take place using structured data analysis processes integrated in Jupyter Notebooks.
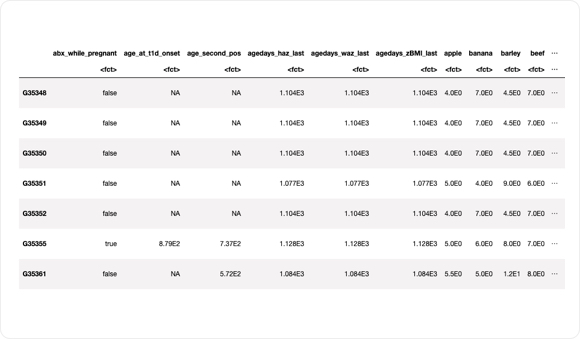
As the table above shows, users can directly sort and access metadata through the semantic framework. This enables users to easily select samples of interest for further downstream analysis.
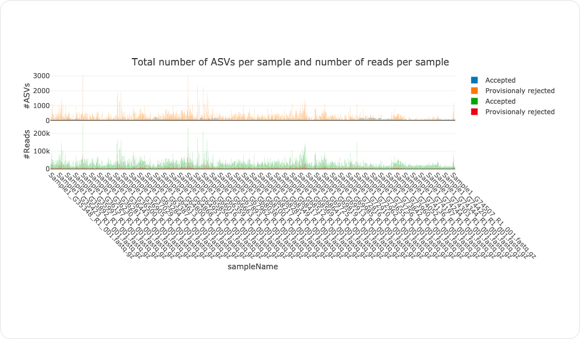
As shown in the graph above, amplicon data is noisy. NG-Tax 2.0, however, suppresses this noise by provisionally rejecting ASVs of low abundance. Moreover, due to the extensive incorporation of metadata and sequence information, users can access and study each (potentially rejected) ASV individually.
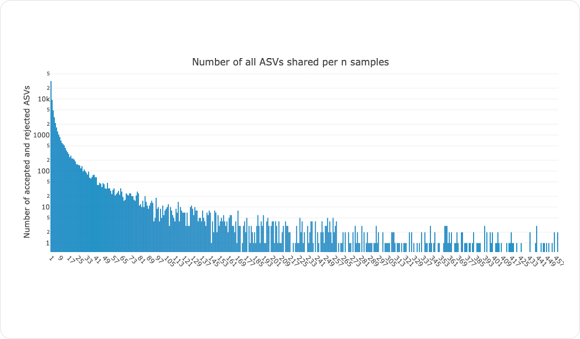
Unlike OTUs obtained by clustering of nearly identical sequences, Amplicon Sequence Variants suppoedly better represent a single species. Since NG-Tax 2.0 extracts ASVs and stores them in a semantic database, it enables cross-mapping of ASVs between thousands of samples. As shown in the grah above, we have plotted the number of ASVs, including the provisionally rejected ASV shared between samples.
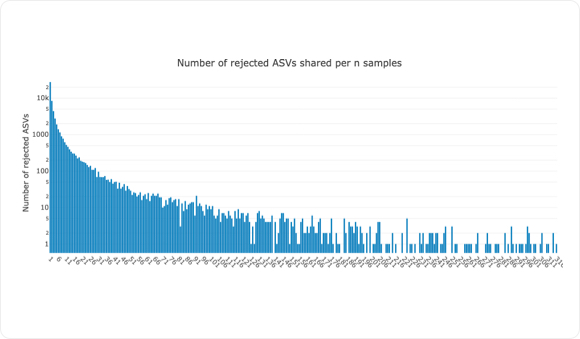
Next, within each sample, NG-Tax analyses and potentially flags ASVs as rejected due to severely low abundant reads, as seen in graph above. These ASVs might be falsely flagged, but through cross-mapping within and between samples these ASVs can become accepted and used for further downstream analysis.

For each individual sample, NG-Tax 2.0 provisionally rejects ASVs of low abundance. While most of these ASVs are likely random noise signals, a provisionally rejected ASV can also point at microbial species with a particular low abundance in the specific sample. We can study this by asking two related questions.
1. “Do we find a particular provisionally rejected ASV in more samples?”
2. “Is this ASV an accepted ASV in other samples?”
As the ASVs are stored in a graph database, this and the results presented in the examples above, can be done with standard SPARQL queries. In the example, we have plotted the number of the provisionally rejected ASV that are shared between the 1800 samples. As can be seen, by far the majority occurs in less than three samples and on the basis of this most probably most of them are noise ASvs. The example SPARQL query above explores this a little bit further and provides an answer to the question “How many provisionally rejected ASVs are in fact accepted ASVs in other samples?”.
