In a three-piece blog series, we introduce the concept of our FAIR Data Station. In the first and second part, we explained how to get started and why. Finally, in this part, we explain how you can build your own semantic database.
By the FAIR Data Platform / November 30, 2021
KEY MESSAGES
- Standardization of metadata, made easy by our FAIR Data Station, ensures that the data is understood in the same way by everyone.
- After acceptance and validation of your Excel file (first blog), a Linked Data file is automatically created.
- These files can be used in applications such as Apache Jena or GraphDB.
The FAIR guiding principles are meant to Find, Access, Interoperate, and Reuse data in an easy way. Our FAIR Data Station guides researchers in metadata management via a simple three-step process, as described in the first blog of this series. Given the fact that many subsidized research projects are required to have their data accessible, reusable and preserved, data FAIRification is obviously important in Life Sciences, You can find additional reasons for being FAIR in the second blog of this series. This third blog concludes with explaining how you can build your own Semantic database, including an example.
Building your own Semantic database
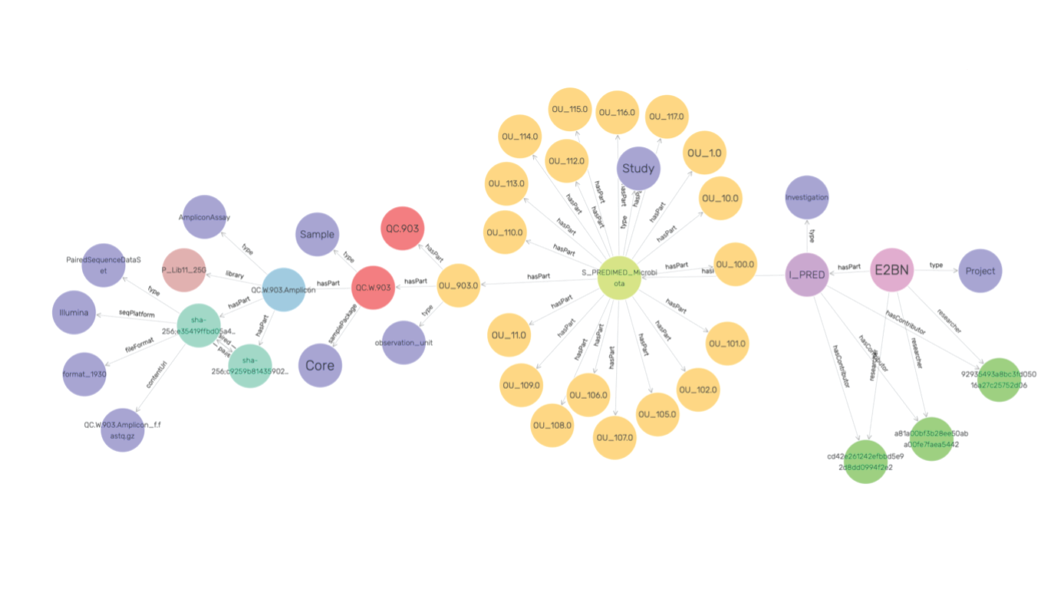
The use of metadata standardization in a community-accepted data format ensures that data elements are understood in the same way by all communicating parties. All in all, this allows for a semantic level of interoperability. After validation and acceptance of your Excel file, a Linked Data file in the TURTLE syntax and file format is automatically created. Next, this file, with file extension .ttl, can be imported into a Semantic Graph Database such as Apache Jena or GraphDB and allows researchers to query project metadata. Additionally, when multiple TURTLE metadata files from neighbouring projects are selected for import, metadata queries across the different projects is possible using the SPARQL query language.
In the example below we queried the metadata of five different studies focusing on microbial biodiversity in soils. To this end, an example SPARQL query listing for each study the number of samples obtained is shown.
PREFIX jerm: <http://jermontology.org/ontology/JERMOntology#> PREFIX schema: <http://schema.org/> select distinct ?study_id (COUNT(?sample) AS ?count) where { ?study a jerm:Study . ?study schema:identifier ?study_id . ?study jerm:hasPart/jerm:hasPart ?sample . } group by ?study_id |
study_id | count |
ERP009103 | 72 |
ERP022327 | 383 |
ERP106967 | 60 |
ERP107112 | 180 |
ERP107808 | 558 |