Last month, researchers from our FAIR Data Platform published a paper on the FAIR Data Station: A platform that helps you to manage your research metadata according to the FAIR principles. In a previous series of blogs, we already explained why FAIR data is so important. In this blogpost, we discuss the main features of the FAIR Data Station and help you get started with FAIRifying your research data.
By the FAIR Data Platform / 13 April 2023
KEY MESSAGES
- The FAIR Data Station aims to helps you to FAIRify your research data
- This online application consists of three modules that help you to create a metadata template, validate this file once filled out, and generate files that allow cross-project searches and publication of sequence data.
- Documentation, including a tutorial, can be found online.
- Go directly to the FAIR Data Station
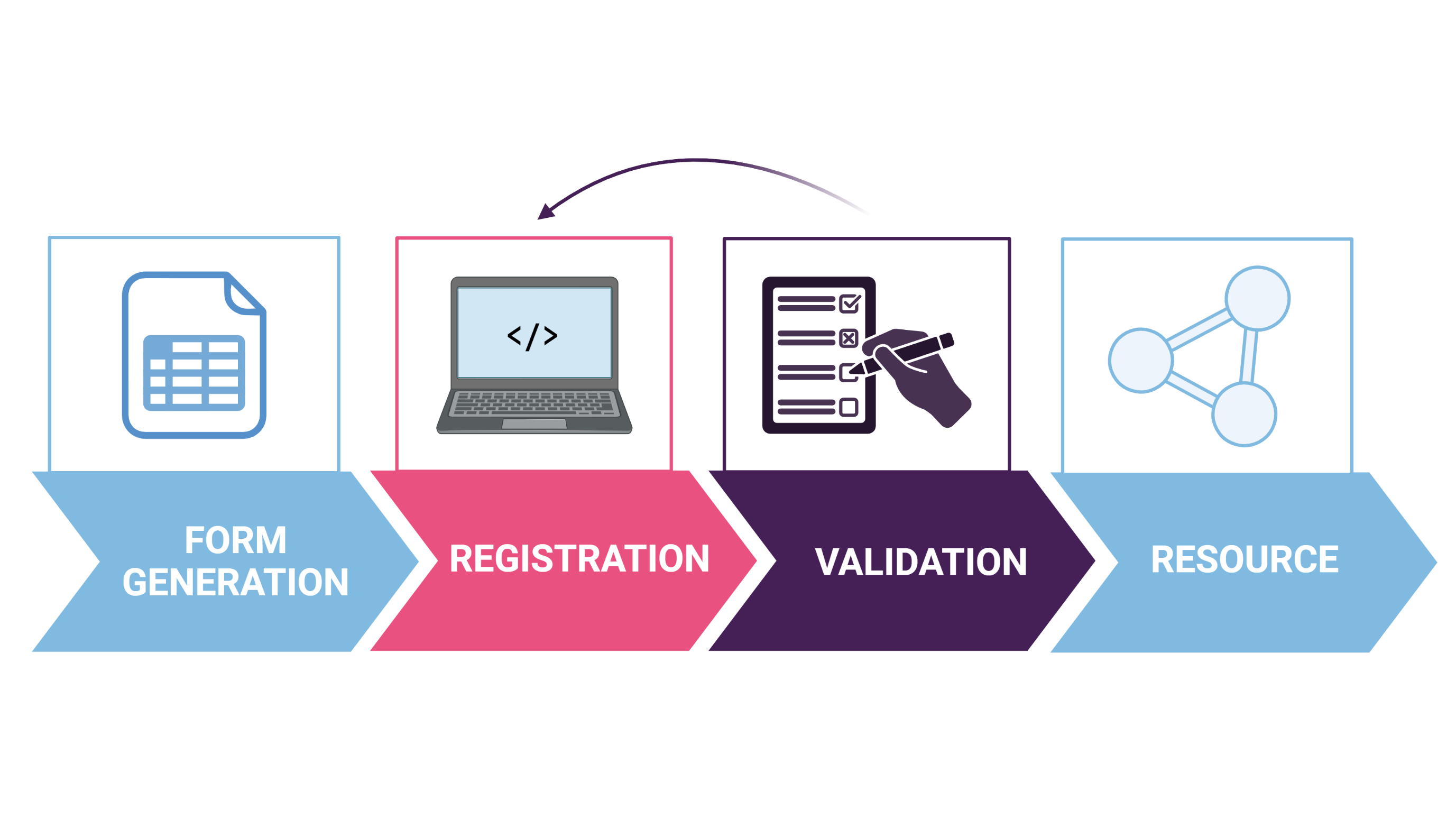
The life sciences are one of the biggest suppliers of scientific data, but without metadata, this data is meaningless. The FAIR Data Station is a lightweight application that aims to support researchers in managing their data according to the FAIR principles. The station consists of 3 modules:
The form generation module creates a metadata template, based on the minimal information model(s) you select.
- The selection of models depends on your experiment (e. the minimally required information describing your samples), and can consist of both mandatory and optional attributes. A list of terms for different experiments can be found here.
- The metadata template generated is an Excel workbook, of which the header rows contains machine-actionable attribute names. Next, you can start registering your sample metadata in this file. The open Excel format allows for offline, on-site metadata registration and supports collaboration and information collection in high throughput.
The validation module allows you to check the format of the recorded values in your metadata file at any time. This is done using a package library of 40 frequently used minimal information checklists. The validation module also checks for activation of unsolicited auto-complete and auto-correction functions and for mismatches between identifiers used at the different ISA levels.
The resource module. After validation of your data, your Excel workbook is automatically exported as a Resource Description Framework (RDF). This document allows you to search through metadata from different programming languages and incorporate the metadata in your analysis workflow. It also allows you to search through multiple research projects that are centred around a common theme. If you want to publish sequence data, you can use the resource module to convert your Excel workbook to an European Nucleotide Archive-compatible XML metadata file.
How to get started?
If you want to get more technical information, please visit the documentation. For instance, you can learn how to set up your own FAIR Data Station and how to modify and extend the existing metadata templates. If you want to start a community to create metadata templates feel free to contact us. If you want to start a community to create metadata templates do not hesitate to contact us too!
If you want to FAIRify your own data, but still do not exactly understand how, you can follow the tutorial, including an explanation of the terms used. If you have questions or suggestions, feel free to contact us. Lastly, if you are an UNLOCK-user, you can also use the FAIR Data Station to access your data files, folders, and data usage.
About the publication
The complete publication on the FAIR Data Station, including relevant links, can be found below:
Lead image created with Biorender.com