The mission of UNLOCK is to unlock microbial potential by looking at whole microbial communities, instead of single species. These communities, however, can be very complex and challenging to investigate. A way to tackle this problem, is to use a simplified, synthetic community of only a selection of key microbes. Recently, UNLOCK-scientists – in collaboration with scientists from the NWO Gravitation program SIAM – followed this approach. They composed a synthetic community of ten key microbes from the human intestinal tract. In this article, we discuss their main findings.
By the Biodiscovery Platform / April 22, 2022
KEY MESSAGES
- The human gut is home to a complex community of microbes which work together, but also compete with each other, for food
- Recently, researchers composed a defined microbial team consisting of ten key gut bacterial species and studied their behavior
- Together, these ten bacterial species interact with each other and convert complex dietary fibers into short chain fatty acids
- This paper strengthens the mission of UNLOCK to unlock microbial potential by looking at whole microbial teams instead of individual bacteria
In the human intestinal tract, dietary fibers that cannot digested by human enzymes are left to the intestinal microbiota residing in the large intestine. Here, the fibers are degraded by a community of microbes, which produce compounds (e.g. simple sugars and short-chain fatty acids) which can be used by other bacteria or human intestinal cells. The interactions between these bacteria is, however, rather complex. Some species co-operate and exchange compounds (called “cross-feeding”), whereas others compete for the same fibers. The latter does not mean that these species cannot live together in the human intestinal tract. Instead, these species have found a way to co-exist, but how they do this exactly remains obscure.
In any case, this mystery cannot be solved by looking at single species or pairs of species only. Clearly, there’s a need to study bacterial species in the context of their community. In a recent study by (former) UNLOCK-scientists, a community of key bacterial species of the human intestinal tract were put together, to unravel their interactions in an environment resembling the human large intestine.
How to design a minimal synthetic community
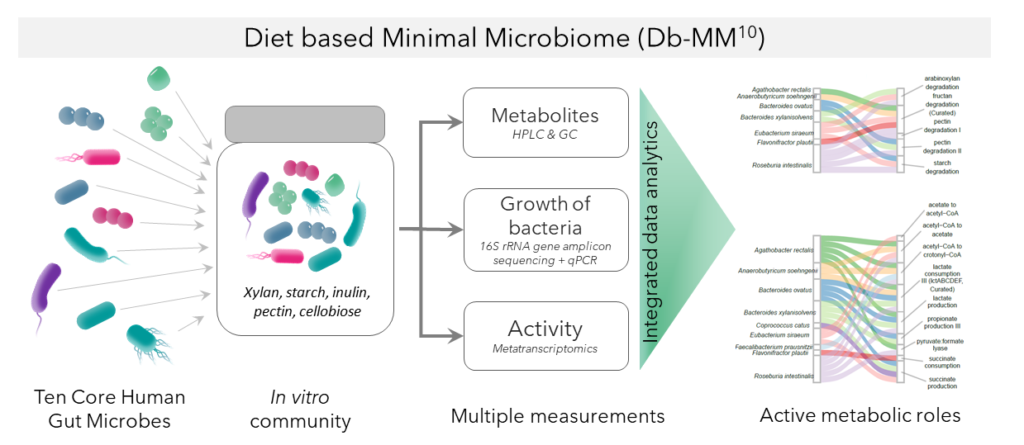
From single members to a whole community
Monitoring the activity of the community
Summary
Read more
If you want to read the full publication, including more detailed information on the bacterial strains and their characteristics, click the link below.
The work described exemplifies the research done at our Biodiscovery Platform.
This study was funded by UNLOCK and the Soehngen Institute of Anaerobic Microbiology (SIAM), and carried out at the Laboratory of Microbiology at Wageningen University & Research.
