Our Parallel Cultivation platform allows the cultivation and investigation of a wide range of microbial communities in extremely well-defined conditions. This article exemplifies the use of the platform with a case study on polymer storage by micro-organisms under different temperatures.
By the Parallel Cultivation Platform / July 28, 2021
KEY MESSAGES
- Studies on microbial communities often lack the link between structure and function
- The Parallel Cultivation Platform unlocks the investigation of microbial communities under different conditions at the same time
- This case study describes the investigation of the effect of temperature on microbial competition between polymer-storing bacteria and "growers"
- In summary, shows the advantage of operating multiple bioreactors in parallel, to answer this research question
The advent of techniques to quantify microbial genomes, has enabled researchers to characterize the composition of microbial communities in a wide range of ecosystems. Research on microbial communities, currently, often lack insight in the functional performance of these communities. Linking community structure to function, however, would greatly improve the understanding of microbial ecosystems and its applications. Indeed, well-controlled bioreactors allow the investigation of both microbial community structure and function, but sampling and analysis of communities can interfere with bioreactor operation. Additionally, differences in study design limit the comparison of datasets from different studies with varying parameters. Clearly, there’s a need for a platform such as the Parallel Cultivation Platform, which allows the investigation of structure and function under different conditions in parallel.
Case study: Linking microbial community dynamics and functionality
To exemplify the use of the Parallel Cultivation Platform, we decribe a recently published study at – a.o. – the Delft University of Technology (TUD, citation below). In this work, we investigated the effect of temperature on microbial competition in parallel-operated reactors. To this end, the mixed microbial community in these bioreactors was pulse-fed with a single nutrient. As shown in previous studies, microbes compete for the limiting nutrient with at least two strategies: The “growers” directly convert the nutrient into biomass, whereas the “hoarders” store the nutrient as internal storage compounds (polymers) and convert them to biomass at a later timepoint.
Polymer storage by bacteria
Hoarders have a competitive advantage over growers, as production of storage polymers is simpler than biomass production and consumes less energy. Therefore, the enrichment of storage polymer-producing bacteria has great industrial potential, especially in the field of resource recovery from waste streams. What kind of storage polymer is formed, depends on the nutrient. In general, waste streams contain a lot of volatile fatty acids, which bacteria convert to polyhydroxyalkanoates (PHA). In turn, PHA can be used in the production of bioplastics and biochemicals. The picture below shows microbes that have stored PHA. Polyhydroxybutyrate (PHB) is the most commonly produced type of PHA by different bacteria.
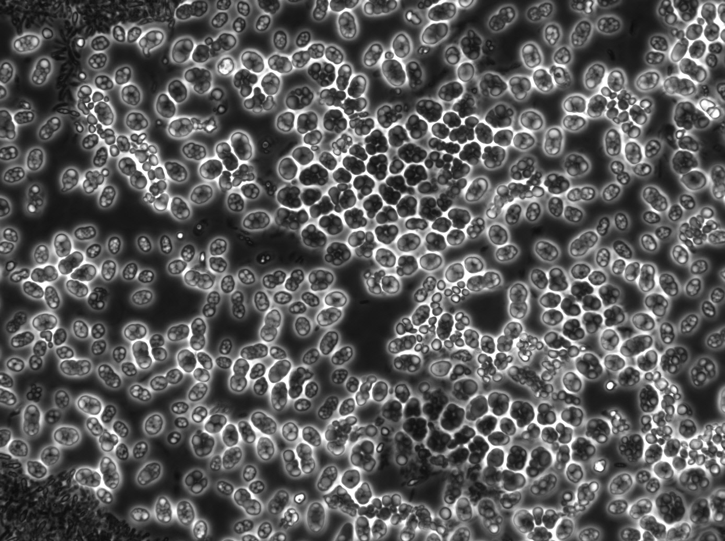
Microscopy picture of microbial cells with PHA inclusion bodies, which are able to store up to 90% of the dry weight as PHA. Figure adopted from Stouten et al. (2019).
Experimental set-up to study polymer storage
To study the effect of temperature on PHB-storing bacteria, eight parallel bioreactors were pulse-fed with acetate and operated in the temperature range of 20°C to 40°C. We operated these sequencing batch bioreactors for over 100 days and monitored them online with state-of-the-art equipment. Amongst others, variables measured included O2 and CO2-off gas, base and acid dose, bioreactor temperature, dissolved oxygen and pH. Subsequently, we used online data to reconstruct functional development of the enrichment cultures, as was verified by offline measurements.
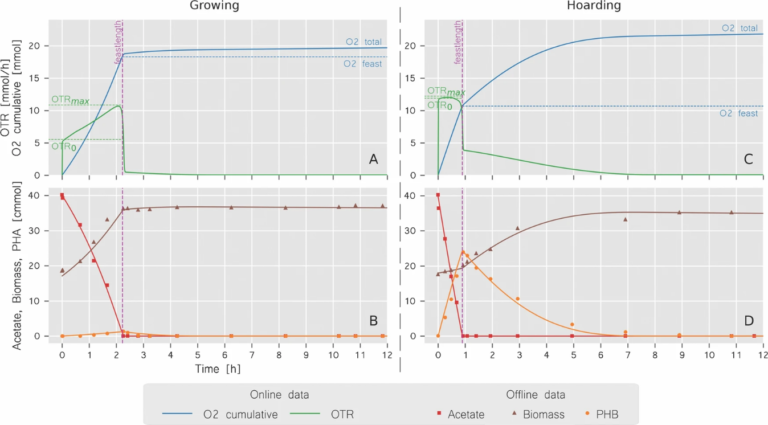
Online and offline data of a single bioreactor cycle, where the online data could be used to a large extent to describe the functional behaviour of the enrichment culture. The gradual enrichment of hoarding biomass in the bioreactors could be monitored over the 200+ consecutive cycles. Figure adopted from Stouten et al. (2019).
Data processing and analyis
Utilizing all online data helped to gain insights in the functional development of the enrichment cultures over time. Moreover, we developed a new data processing pipeline to improve data handling and data interpretation by converting online measurements to real time microbial activity. To this end, for each operational cycle of each bioreactor, the microbial functionality was captured as a mathematic depiction of the microbial activity. Eventually, three characteristic indicators were derived that showed strong correlations with microbial community structure development and offline-verified measurements, as reflected in the figure below for one bioreactor system.
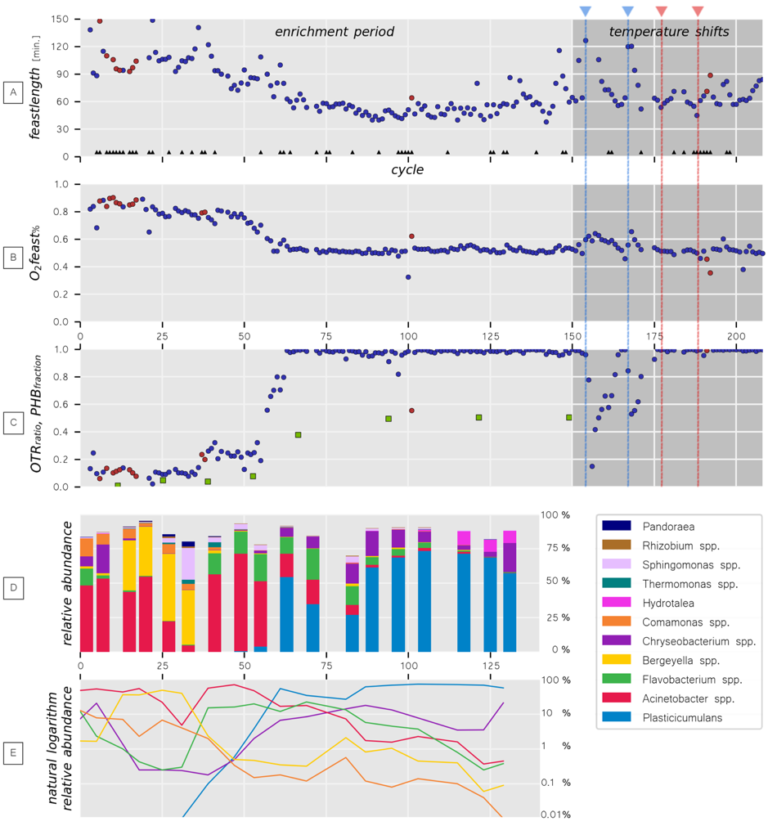
Detailed overview of the output of the Parallel Cultivation Platform at TUD, which is shown to exemplify the conceptual set-up of the different reactor types included in UNLOCK. Figure adopted from Stouten et al. (2019).
Major benefits for this case study
In summary, major benefits of the UNLOCK Parallel Cultivation Platform for this research included the ability to operate eight bioreactors in parallel from the same inoculum at high resolution. All in al, the online data processing tools, combined with the community structure analysis by 16s rRNA gene amplicon sequencing allowed for a novel experimental design and aided in the understanding of competitive strategies in microbial communities.
Interesting links
If you got inspired, read more about our Parallel Cultivation Platform.
