Our Biodiscovery Platform enables the isolation and investigation of yet unknown microorganisms. Subsequently, these microbes can be used as building blocks for synthetic microbial communities. In this article, we describe the design of a synthetic microbial community from a source of microbes we always carry with us: the human gut.
By the Biodiscovery Platform / July 14, 2021
KEY MESSAGES
- The understanding and application of synthetic microbial communities become increasingly popular. It can, however, be challenging.
- By combining the different expertises within our UNLOCK team, we can accelerate the design and characterization of synthetic communities.
- Eventually, the design of a synthetic community representing the human gut microbiome will lead to better understanding. Ultimately, this may help to improve human health.
The role of UNLOCK
A synthetic gut microbial community - a case study
Approach
First, we screened over 5000 shotgun metagenomics datasets from public databases to identify highly prevalent core bacterial species in the human gut. Next, we shortlisted 10 core gut bacteria. We did this by using a combination of genome-based prediction; in silico metabolic network-based predictions of competition and complementarity; and the available information on the physiology of core bacteria. In brief, the design was aimed towards efforts to
- Firstly, reconstruct the central metabolic pathway from diet to SCFA production,
- Secondly, identify the metabolic basis for co-existence of competing core species,
- Lastly, identify contributions of each microbial strain to multiple functions related to degradation of carbohydrates and production of SCFA
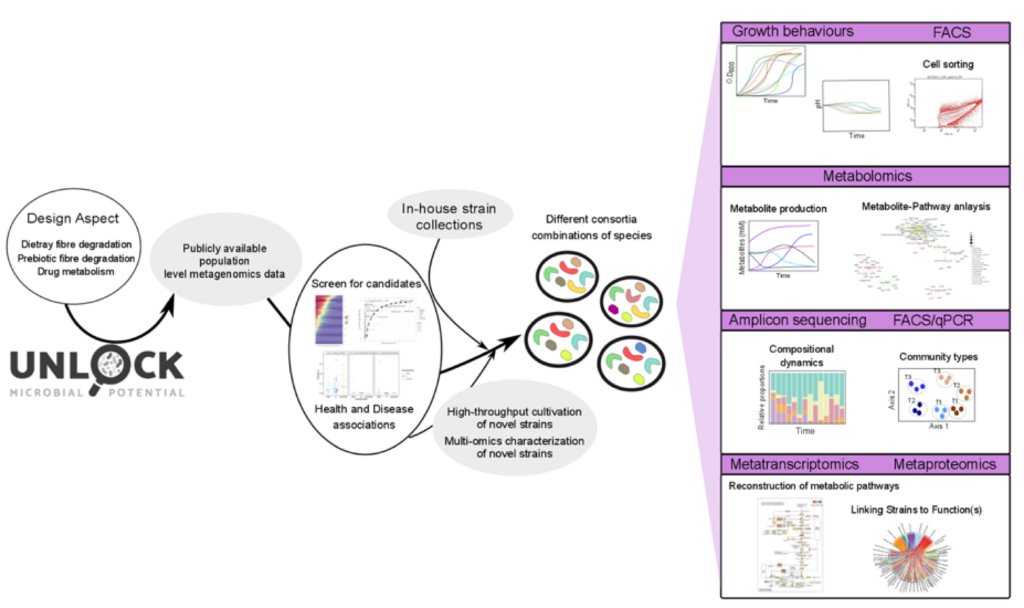
An overview of the process from datamining to design and investigation of synthetic gut microbiomes, which helps unravelling community stability and function. Figure by Sudarshan Shetty.
Integration of the FAIR Data Platform
In summary, we carried out metabolite measurements, quantitative microbiota profiling and metatranscriptomics. Subsequently, three-way integration and identification of key ecological and functional aspects of the synthetic gut microbiome can be achieved through our FAIR Data Platform. This platform comprises bioinformatics pipelines, which process raw data and generate output for species abundances as well as active gene/pathway abundances. This output can be further analysed together with metabolite data using our custom workflows provided as Jupyter Notebooks. For the whole workflow, see the figure above.
Interesting links
If you got inspired, read more about our Biodiscovery Platform.
When you are interested in synthetic and minimal communities as models for the human gut microbiome, read this publication by a.o. Hauke Smidt, manager of the Biodiscovery Platform:
